Gut microbiota is essential for a healthy fish. It can be influenced by genetic background of the fish, novel feed ingredients, and it can be stimulated by feed additives. But how can we quantify the composition of gut microbiota community in fish?
Gut microbiota is elemental for fish health
The composition of fish gut microbiota is of great importance for the host’s physiology, immunity and the use of feed. A comprehensive view of fish intestinal microbiota, including both taxonomic and genetic composition, is highly relevant for aquaculture practices, especially when novel ingredients are used in feeds.
But how to study the diverse gut microbiota in fish? Decades ago, the initial trials were conducted by sampling the gut and culturing the microorganisms in cell cultures in a laboratory. But this resulted in poor success and biased cell replication under culture conditions, causing artificial shifts in the community composition and function.
DNA-based methods for quantifying gut microbiota
Only when the novel DNA-based methods were developed, accurate evaluation was possible and no cell cultures were needed.
The DNA based methods include:
- Quantitative real-time PCR (qPCR), used for quantitative analysis of taxa.
- Clone libraries for identification of microbiota composition.
- Finger-printing methods such as temporal temperature gradient electrophoresis (TTGE) and denaturing gradient gel electrophoresis (DGGE).
- Fluorescent in situ hybridization (FISH) used to determine the abundance of particular taxa, total microbial levels and assess bacterial–host interactions at the mucosal brush.
- High-throughput, next-generation sequencing (NGS).
NGS has been the latest introduced technology to study the composition and genetic potential of complex microbial communities, such as gut microbiota. The most sophisticated NGS platforms generate large volumes of DNA sequencing data in a single run, enabling the rapid and cost-effective acquisition of in-depth and accurate sequence data and allowing for the detection of both dominant and low abundance (rare) microbial community members.
Variants of 16S rRNA gene identify members of microbiota
Due to its universal phylogenetic distribution across taxa, the gene coding 16S rRNA has been one of the most important targets of NGS applications for investigating the bacterial communities in fish gut (Fig. 1).
16S rRNA gene contains fast-evolving regions (hypervariable regions V1-V9) that are used to taxonomically classify organisms up to genus level, while species-level resolution might be unfeasible using this information alone. Another advantage is that there are several large databases of already analysed species and genus to which the yet unidentified 16s rRNA results can be compared to.
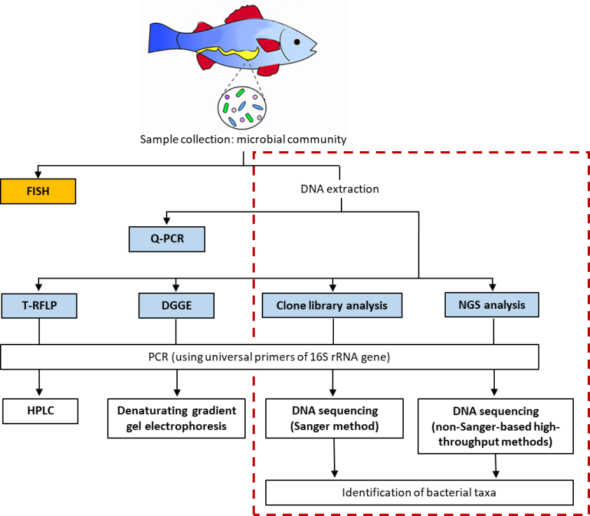
Figure 1. Workflow of DNA molecular methods targeting 16S rRNA gene. The broken line encircles the methods based on sequencing technologies (Modified from Source 1; Creative Commons License).
In recent years, there has been very rapid advancement in sequencing technology (increased coverage and deep sequencing) and bioinformatic tools which have improved the accuracy of the DNA-based methods.
AquaIMPACT approach
AquaIMPACT uses NGS techniques based on the use of the Illumina platform. Alignments are performed by establishing high stringency filters (≥ 90) of sequence identity and query coverage, and the resulting assignation results are filtered and data summarized in Operational Taxonomic Units (OTUs). The used methods and filters reduce the core microbiota to 200-300 OTUs that can be used to identify the representation of the Phylum Proteobacteria, Firmicutes and Actinobacteria in the gut of fish with different age, sex, and nutritional and genetic background.
Expected AquaIMPACT outcome
The expected outcome is to establish the connection between individual microbiota and the corresponding metabolic fish phenotype that leads to the superior fish performance and health. In other words, which microbiota community generates vital farmed European seabass and gilthead seabream (Fig. 2)?
Figure 2. Gut microbiota colonization. Which gut microbiota community generates vital fish?
The AquaIMPACT trials are currently conducted at the ULPGC (University of Las Palmas in Gran Canaria) facilities with Skretting ARC diets, being the microbiota analysis conducted by USI (University of Insubria) for European seabass, and CSIC (Consejo Superior de Investigaciones Científicas) for gilthead seabream.
References
1 – Butt RL and Volkoff H (2019). Gut microbiota and energy homeostasis in fish. Front. Endocrinol. 10:9. doi: 10.3389/fendo.2019.00009
Authors
Genciana Terova, Simona Rimoldi. University of Insubria (USI), Italy; Jaume Pérez-Sánchez, Fernando Naya, Carla Piazzon, Ariadna Sitjà-Bobadilla. Consejo Superior de Investigaciones Científicas (CSIC) Spain.
